 IsotopeR is a mixture model framework designed for stable isotope analysis in R. IsotopeR uses primarily stable isotope data for sources (foods) and mixtures (consumers) to predict the proportional dietary contributions of consumers. Although other biochemical tracers can be used within the IsotopeR framework, our modelling approach was designed to carry out diet reconstructions using stable isotope data and other relevant information (see User Guide & example data). We intend to continue to make IsotopeR’s graphical user interface (GUI) in R simple and intuitive and welcome any feedback.
IsotopeR is a mixture model framework designed for stable isotope analysis in R. IsotopeR uses primarily stable isotope data for sources (foods) and mixtures (consumers) to predict the proportional dietary contributions of consumers. Although other biochemical tracers can be used within the IsotopeR framework, our modelling approach was designed to carry out diet reconstructions using stable isotope data and other relevant information (see User Guide & example data). We intend to continue to make IsotopeR’s graphical user interface (GUI) in R simple and intuitive and welcome any feedback.
IsotopeR currently includes a number of features, giving researchers analytical flexibility. Some of these features currently include:
- A hierarchical Bayesian design
- Proportional dietary contributions predicted for the population, groups (e.g., sexes), and individuals
- Corrections to the isotopic mixing space for the effects of food stoichiometry and differential digestibility of elements
- Unlimited stable isotope systems
- > n+1 sources (n = stable isotope systems)
- Includes process and measurement errors
- 2-D and 3-D (rotational) mixing space plots
- DIC estimation for use in model selection
- User-friendly GUI
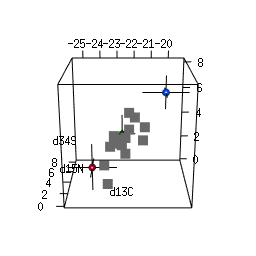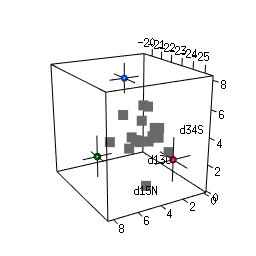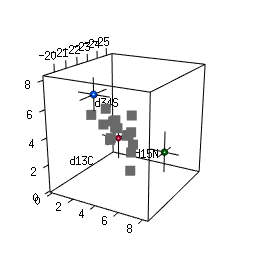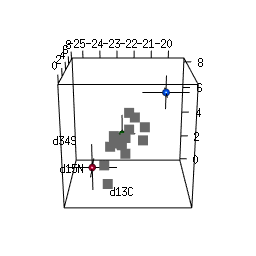 IsotopeR installation and operation
IsotopeR installation and operation
- Download the most current version of R
- Install JAGS for Mac or Windows.
- Mac users with R before v3.0 must install tcltk and all mac users must also install XQuartz 2.7.8. For smoother operation, update your computer to R 3.2.2 for Mac OS X 10.9 or higher.
- Install “IsotopeR” and the following dependencies (packages: fgui, runjags, ellipse, plotrix,
colorspace) using the following code in R:
> install.packages(“IsotopeR”, dep=T) - Open our User Guide and run through our examples using our example data
- If you have problems contact us with questions
Cite IsotopeR package
Ferguson, J.M., & J.B. Hopkins III. IsotopeR. 2015. IsotopeR: Stable Isotope Mixing Model. R package version 0.5.1.
Modeling papers and details about IsotopeR
Hopkins, J.B. III, & J.M. Ferguson. 2012. Estimating the diets of animals using stable isotopes and a comprehensive Bayesian mixing model. PLoS ONE 7: e28478.
Hopkins, J.B. III, J.M. Ferguson, D. Tyers, & C.M. Kurle. Selecting the best stable isotope mixing model to estimate grizzly bear diets in the Greater Yellowstone Ecosystem. PLoS ONE 12(5): e0174903.
Other tools
Posterior overlap and probability of similarity for comparing proportional dietary estimates to determine if distributions are similar.

